Our Project
Background and Motivation
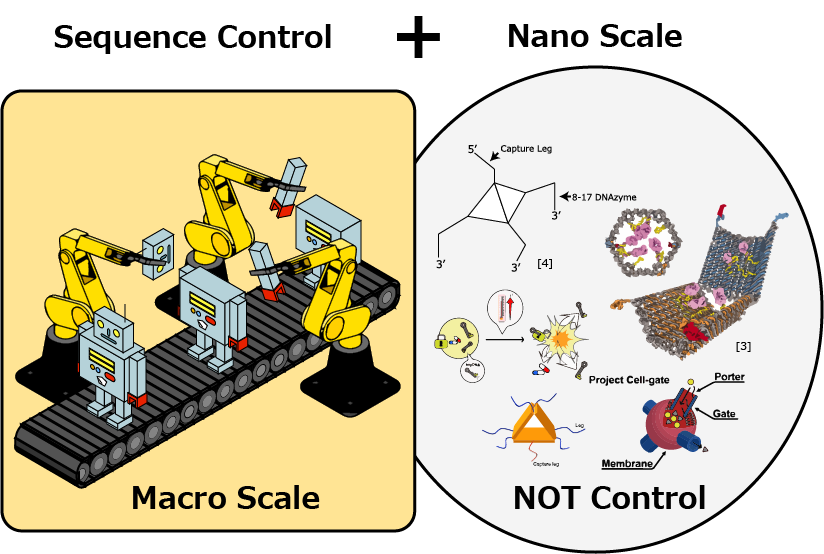
Developing molecular devices and systems is one of the goals of nanoengineering. One approach to the goal is using DNA, whose reactions can be programmed based on the Watson-Crick base pairing. By using DNA, a variety of nanodevices have been developed so far [1~6]. Orchestrating different nano devices in a consistent manner is a challenging topic in nano-enineering. In macroscale, this kind of problem is ubiquitously found in factory automation. Here, we focus on "sequence control", which is a technique used in control engineering. Sequence control gives us a powerful method to synchronize the workflow of devices and machines. It controls them by giving commands in order. Applying this method in nanoscale, we are able to open a way to control molecular devices in a sequential manner.
Project Idea

To realize this concept, we have designed a device that generates single stranded DNAs in desired order. In this project, we took two different approaches. One approach is to achieve the function by a combination of DNA and enzymes (Enzymatic device). The other approach is using only DNA (Enzyme-free device). Depending on the specific purpose, one can choose one of the devices.
In principle, both of them are universal. Namely, there are no limitation in number and permutation of outputs. We analyze the behaviors of our devices both in vitro and in silico to demonstrate their functionality.
Project Goal
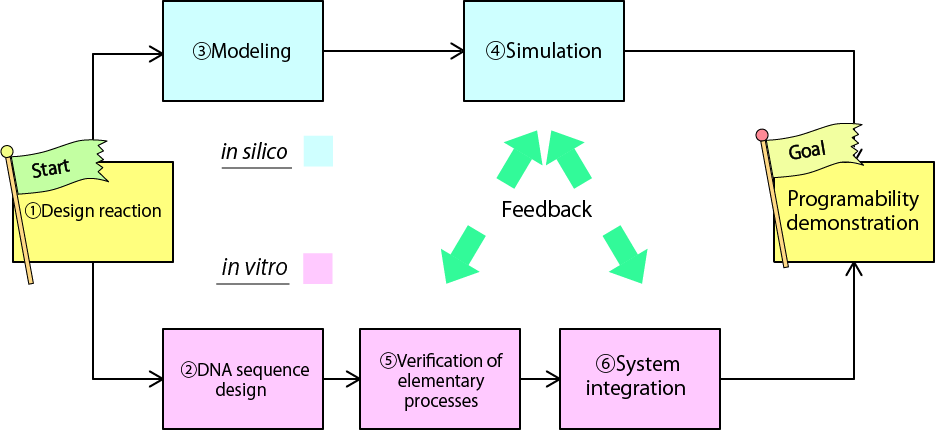
The goal of our project is to realize the devices both in vitro and in silico. We have divided our goal into 6 sub-goals.
Achievement
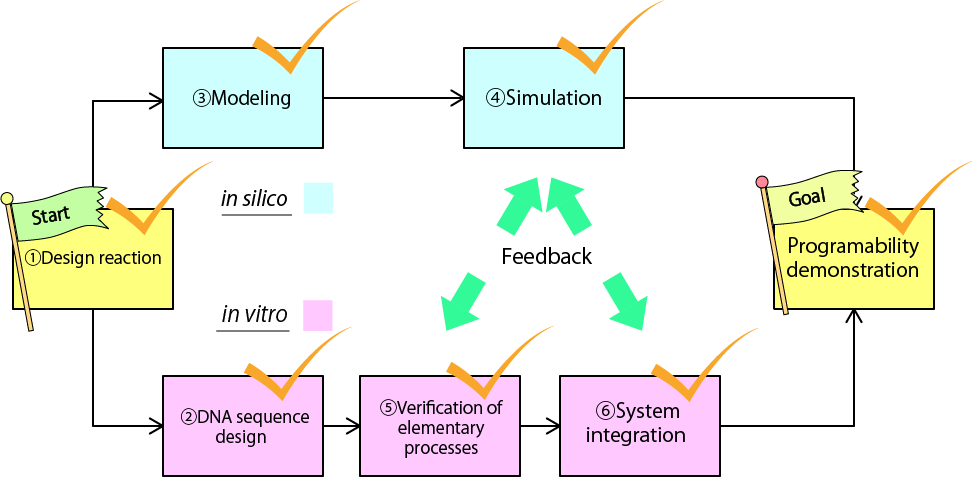
Actually, we have achieved all the sub-goals so far. Our devices behave almost perfectly as designed both in vitro and in silico. We can program the number and permutation of output releasing. In addition, the time intervals between outputs are also programmable by changing concentrations of DNA species.
Future
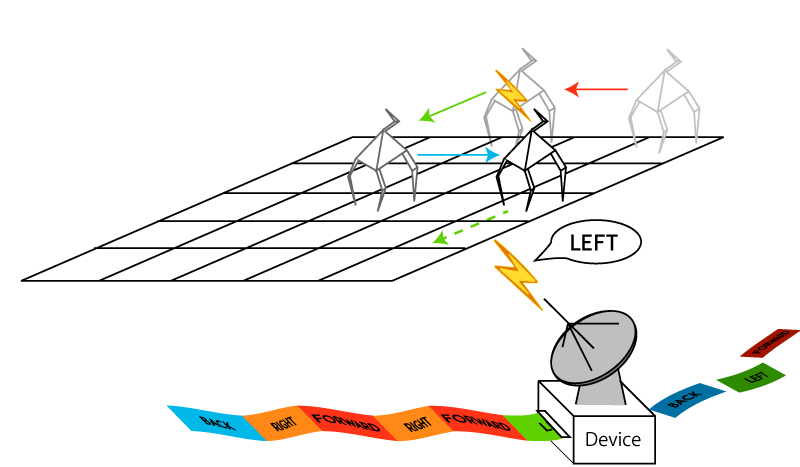
Our devices enable us to control variety of nanoscale machines in a sequential manner. For example, we can control molecular walkers [3,4] giving them motion instructions by using our device.

Another application is autonomous reconfiguration of DNA nano structures. In macroscale, self-reconfigurable structures which are capable of transforming into arbitrary shape have been developed [7]. Our device can realize it in nanoscale. By coding instructions to assemble DNA structures on the input tape, rearrangement of nano-modules will be realized automatically.
